Bovine viral diarrhea virus (BVDV), also known as Pestivirus bovis, is one of the most important viruses affecting bovine species worldwide. BVDV belongs to the genus Pestivirus in the family Flaviviridae, which includes other viruses affecting sheep and swine species, namely border disease virus (BDV) and classical swine fever virus (CSFV). These viruses cause significant economic losses to the global livestock industry.
Bovine viral diarrhea virus (BVDV) causes immunosuppression, leading to Bovine Viral Diarrhea (BVD), also known as mucosal disease, with a wide range of clinical signs. This disease results in significant economic losses and is found in the majority of countries worldwide.
BVDV is a positive-sense single-stranded RNA enveloped virus with a 12.5 kb genome. It encodes a single polyprotein that is post-translationally cleaved by viral and cellular proteases into four structural proteins (C, Erns, E1, and E2) and eight nonstructural proteins (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A, NS5B).
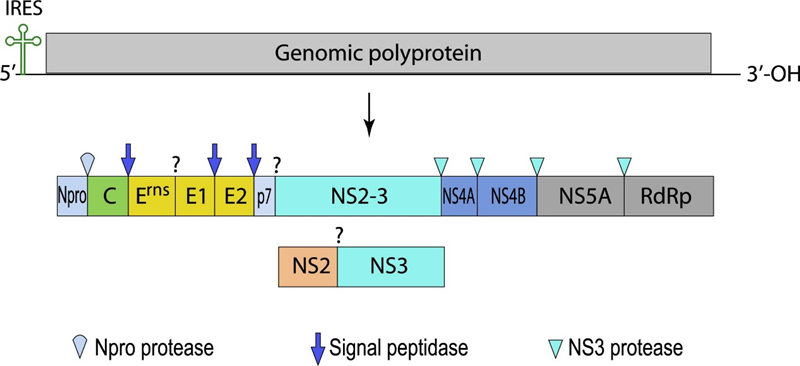
Fig 1. Genome organization and schematic representation of BVDV (viralzone)
Npro
Npro is a leader cysteine autoprotease that cleaves itself from the nascent polyproteins to produce the C protein. The active center of Npro contains two critical amino acids, Cys69 and His130. Unlike other viral proteins, Npro has no counterpart in the other members of the Flaviviridae family. Npro can suppress the transcription of IFN genes by interacting with IRF7 and S100A9, which severely interferes host’s innate and adaptive immune system. The first 19 amino acids are not essential for its protease activity. The remaining amino acids forms a unique clam shell’s-like fold divided into a catalytic domain and a zinc binding domain.
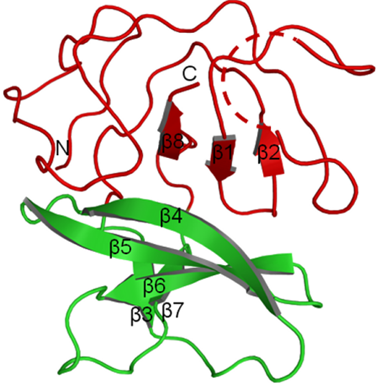
Fig 2. Crystal structure of Npro. The catalytic domain is shown in red and the zinc-binding domain in green (Gottipati et al., 2013)
C Protein (Capsid)
The capsid (C), the most abundant protein, coats the genomic RNA, protect it within the viral particle. Encoded downstream of Npro, the C protein's N-terminal region arises from the autoproteolytic cleavage of Npro. The C protein does not form a higher-order conformation, instead, it interacts with the genomic RNA through electrostatic interaction.
Erns
Erns is the abbreviation of envelope protein RNase secreted. Erns is unique to the Pestivirus genus because it has no counterpart in other members of the Flaviviridae family. Erns can bind with nucleotide substrates and exhibits ribonuclease activity. It is predicted to belong to the T2 ribonuclease family. The Erns protein is a heavily glycosylated protein that contains an amphipathic helix embedded in the membrane. After infection, Erns is released from infected cells and interferes with the immune response.
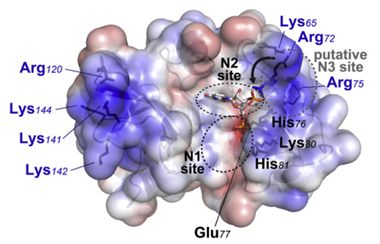
Fig 3. Electrostatic Surface of Erns (Krey et al., 2012)
Envelope glycoprotein (E1, E2)
E2 is the most abundant surface protein of BVDV, and it is covalently linked to E1 by disulfide bonds. The formation of the E1-E2 heterodimer is essential for viral infection. As the largest glycoprotein, E2 is the primary antigenic target for antibodies. However, convalescent cattle serum does not contain significant levels of antibodies against E1. E2 is the most easily recognized antigen by the immune system (immunodominant) and induces the production of neutralizing antibodies in the host following infection or vaccination with live or killed vaccines. The formation of heterodimer of E1 and E2 is ensstial for infection of the virus.
The function of E1 is not well understood, but in comparison to other species in the Flaviviridae family, E1 may function as a chaperone for the E2 protein, preventing its premature activation until infection.
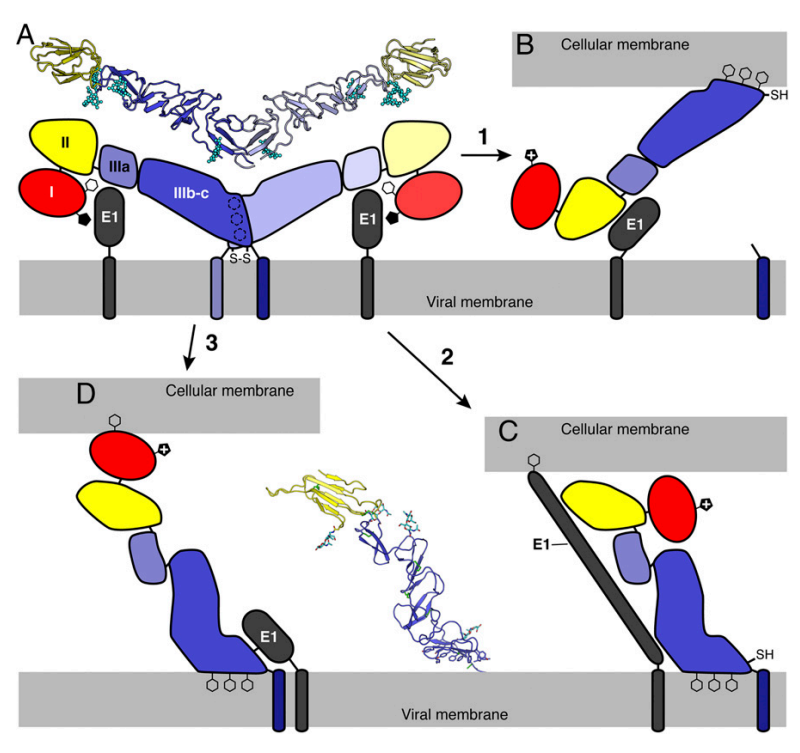
Fig 4. Putative Pestivirus membrane fusion mechanisms (Li et al., 2013)
p7
p7 can regulate the cleavage of E2-p7 by interacting with NS2 and E2, thereby influencing virus production. Additionally, p7 primarily functions as an ion channel.
NS2
NS2 is a cysteine protease with a highly hydrophobic N-terminal region. The C-terminal region is protease domain. The release of NS2 from polyprotein peptide is mediated by NS2 self-cleavage. During infection, DNAJC14 forms a complex with NS2-NS3 to promote the activation of NS2.
NS3
NS3, which has protease, helicase, and NTPase activities, is responsible for cleaving the NS proteins downstream from and including its own C-terminus. The cleavages of NS4A, NS4B, and NS5A are catalyzed by the serine protease domain of NS3, which requires NS4A as a cofactor for full proteolytic activity, forming the NS3-4A protease.
NS4A
NS4A functions as a cofactor of NS3, interacting and forming a complex that catalyzes the cleavage of downstream proteins NS4B, NS5A, and NS5B.
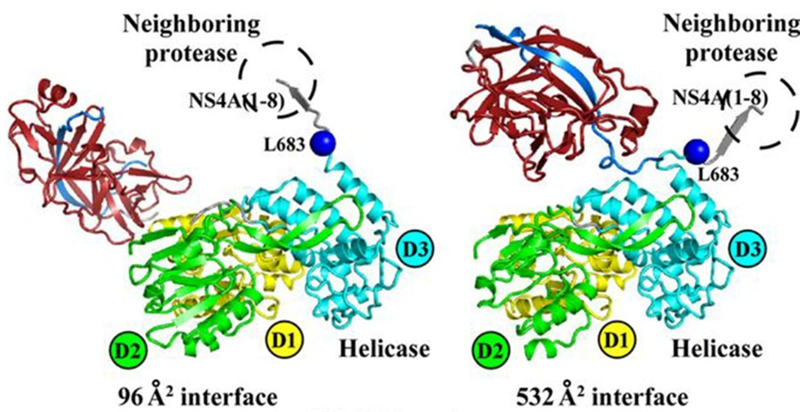
Fig 5. Crystal structure of NS3/NS4A complex (Zheng et al., 2017)
NS4B
NS4B exhibits NTPase activity and participates in BVDV genomic replication. NS4B can induce the formation of autophagosomes to enhance intracellular replication. BVDV can evade the host immune response and establish a persistent infection by inhibiting the innate immune responses of cattle. This inhibition is mediated through the interactions of the viral proteins Npro, Erns, and NS4B with host immune signaling pathways. NS4B interacts with MDA5 to inhibit the RLR signaling pathway and the production of IFN-β, resulting in the suppression of the innate immune response and enabling immune escape. NS4B is a primary target for disease diagnosis and vaccine development.
NS5A
NS5A is a phosphorylated protein which can inhibit the activation of NF-κB signal pathway. NS5A is a component of viral replicase.
RNA-dependent RNA polymerase (RdRp, NS5B)
NS5B is an RNA-dependent RNA polymerase. GTP can stimulate the RdRp activity of NS5B.
Reference
1.Al-Kubati, A.A.G., Hussen, J., Kandeel, M., Al-Mubarak, A.I.A., and Hemida, M.G. (2021). Recent Advances on the Bovine Viral Diarrhea Virus Molecular Pathogenesis, Immune Response, and Vaccines Development. Front Vet Sci 8, 665128.
2.Chase, C.C. (2013). The impact of BVDV infection on adaptive immunity. Biologicals 41, 52-60.
3.Chi, S., Chen, S., Jia, W., He, Y., Ren, L., and Wang, X. (2022). Non-structural proteins of bovine viral diarrhea virus. Virus Genes 58, 491-500.
4.Gottipati, K., Ruggli, N., Gerber, M., Tratschin, J.D., Benning, M., Bellamy, H., and Choi, K.H. (2013). The structure of classical swine fever virus N(pro): a novel cysteine Autoprotease and zinc-binding protein involved in subversion of type I interferon induction. PLoS Pathog 9, e1003704.
5.Krey, T., Bontems, F., Vonrhein, C., Vaney, M.C., Bricogne, G., Rumenapf, T., and Rey, F.A. (2012). Crystal structure of the pestivirus envelope glycoprotein E(rns) and mechanistic analysis of its ribonuclease activity. Structure 20, 862-873.
6.Li, Y., Wang, J., Kanai, R., and Modis, Y. (2013). Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proceedings of the National Academy of Sciences 110, 6805-6810.
7.Neill, J.D. (2013). Molecular biology of bovine viral diarrhea virus. Biologicals 41, 2-7.
8.Peterhans, E., Bachofen, C., Stalder, H., and Schweizer, M. (2010). Cytopathic bovine viral diarrhea viruses (BVDV): emerging pestiviruses doomed to extinction. Vet Res 41, 44.
9.Zheng, F., Lu, G., Li, L., Gong, P., and Pan, Z. (2017). Uncoupling of Protease tran-Cleavage and Helicase Activities in Pestivirus NS3. Journal of Virology 91, 10.1128/jvi.01094-01017.
Applications: ELISA, Immunogen, SDS-PAGE, WB
Expression system: E. coli
Accession: QZX63172.1
Protein length: Glu3-Ser339
Applications: ELISA, Immunogen, SDS-PAGE, WB
Expression system: E. coli
Accession: P19711
Protein length: Met1-Cys168
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: P19711
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: P19711
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: P19711
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: P19711
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: P19711
Applications: ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Expression system: Insect Cells
Accession: Q65808
Protein length: His36-His370
Applications: ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Expression system: Mammalian cells
Accession: AGC12054.1
Protein length: Cys1-Asp218
Applications: ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Expression system: E. coli
Accession: P19711
Protein length: Ala498-Thr663