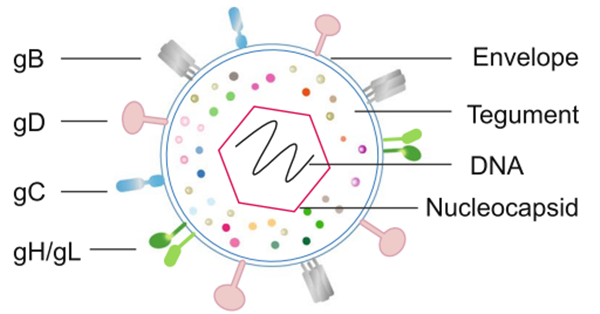
Fig. 1 Schematic representation of HSV (Bai et al., 2024)
Herpes Simplex Virus (HSV) is an enveloped virus belonging to the Herpesviridae family, with two main species: HSV-1 and HSV-2. HSV-1 primarily causes orofacial and genital infections, while HSV-2 is mainly associated with genital infections and severe neonatal infections. HSV infections are widespread, with HSV-1 affecting over 3.7 billion people globally and HSV-2 infecting approximately 500 million people. Once infected, individuals harbor the virus for life, and it can be particularly dangerous in newborns and immunocompromised individuals.
HSV contains a large, linear double-stranded DNA genome, approximately 150 kilobase pairs (kbp) in size. This genome encodes around 280 open reading frames (ORFs), producing about 200 proteins. The viral genome is enclosed with an icosahedral capsid composed of 162 capsomeres. This capsid is surrounded by a protein-rich layer called the tegument, which plays a crucial role in viral replication and immune evasion. The virion is enveloped in a lipid bilayer membrane that includes at least 20 viral proteins, 13 of which are glycoproteins essential for viral attachment and entry into host cells.
The viral capsid is composed with five capsid protein, UL6, UL18, UL35, UL38 and the major capsid protein UL19. Proteins associated with viral replication and immune evasion in the tegument are VP16, UL36, and VP22. The viral envelope glycoproteins, including glycoproteins B, C, D, and E, are critical for the virus’s ability to invade host cells and evade the host immune response.
Despite the existence of antiviral treatments, there is currently no cure or effective vaccine for HSV, underscoring the need for continued research into preventive and therapeutic measures.
gB (glycoprotein B)
Glycoprotein B (gB) is the fusogen of HSV, a type I membrane protein anchored in the viral envelope, and one of the most conserved glycoproteins across all herpesvirus subfamilies. Membrane fusion triggered by gB is highly regulated, requiring the viral glycoproteins gD, gH, and gL, which form the minimal "fusion machine" complex. Glycoprotein K (gK) and other viral components further modulate this process. gB is essential for virion attachment and entry into various cell types. During entry, gB undergoes conformational changes from its pre-fusion to post-fusion state, exposing hydrophobic peptides that embed into opposing membranes to initiate fusion. This transition is regulated by glycoprotein H (gH).
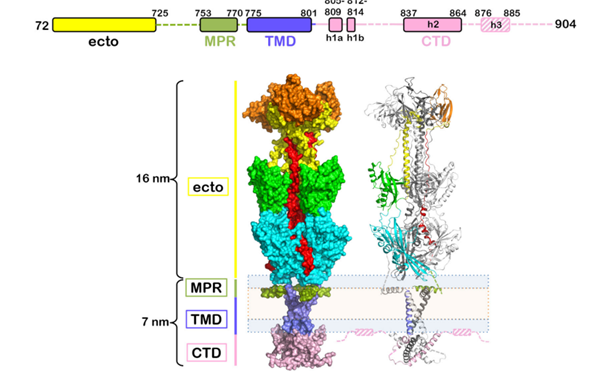
Fig. 2 Domain organization and tertiary structure of HSV-1 gB (Cooper et al., 2018)
gC (glycoprotein C)
Glycoprotein C (gC) is encoded by the UL44 gene, which is conserved across all three alphaherpesviruses: HSV-1, HSV-2, and VZV. gC plays a crucial role in viral entry by binding to cell surface heparan sulfate proteoglycans, serving as the principal viral ligand for this interaction. gC also binds to the complement component C3b, inhibiting complement-mediated immunity and aiding in immune evasion. In human epidermal keratinocytes, gC is one of the primary targets for CD4 T-lymphocyte cytotoxicity. A recent study suggests that HSV-1 gC protects other viral envelope glycoproteins essential for entry, such as gB, by shielding them from neutralization, thereby enhancing viral survival and infectivity.
gD (glycoprotein D)
Glycoprotein gD plays a crucial role in HSV entry by binding to specific cellular receptors, which triggers a cascade of events activating the fusion process. There are at least three known cellular receptors for gD: herpesvirus entry mediator (HVEM), nectin-1, and 3-O-sulfonated derivatives of heparan sulfate. The C-terminal portion of the gD ectodomain is particularly important for HSV-1 infectivity as it influences gD's binding to these receptors, especially nectin-1, making gD itself a key target for neutralizing antibodies. This interaction is essential for initiating the fusion cascade involving gB and the gH/gL complex. The gC interaction with heparan sulfate proteoglycans (HSPG) is important but non-essential for HSV entry, whereas the gD/receptor interaction is critical and considered a prerequisite for subsequent viral entry steps.
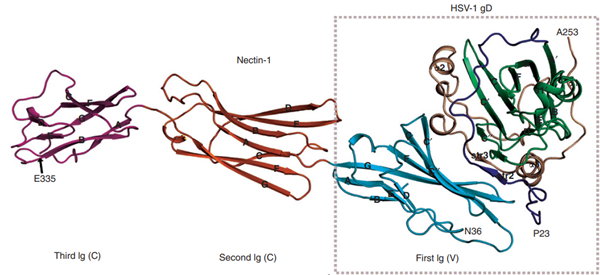
Fig. 3 Structure of gD/Nectin-1 dimer (Zhang et al., 2011)
gE (Glycoprotein E)
The HSV envelope glycoproteins E and I (gE and gI) of both HSV-1 and HSV-2 form a complex that functions as a receptor for IgG to escape detection by the host’s immune system. The regions responsible for IgG Fc binding were mapped to amino acids 235-380 on gE and 128-145 on gI. The binding of monomeric IgG requires the formation of the gE/gI complex, which is responsible for the higher affinity binding of IgG. When expressed alone, gE functions as a low-affinity Fc receptor, binding IgG aggregates, but not IgG monomers. gE-gI binds immunoglobulin G at the basic pH of the cell surface and releases it at the acidic pH of lysosomes, consistent with a role in facilitating the degradation of antiviral antibodies.
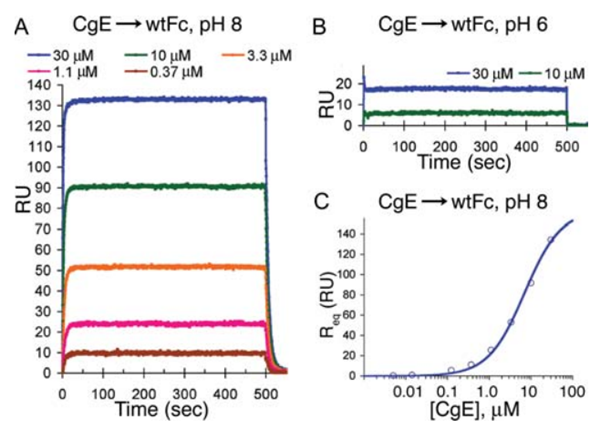
Fig. 4 Binding of gE to Fc at pH 8 and pH 6 (Sprague et al., 2006)
gH-gL complex
gH-gL complex which is found in the virion envelope is essential for virus infectivity and is a major antigen for the host immune system. Polyclonal antibodies to the complex did block entry even when added after virus attachment. These antibodies exhibited high titers of complement-independent neutralizing activity against HSV. gH-gL-immunized mice exhibited high titers of virus neutralizing activity. These data suggest that gH-gL is biologically active and may be a candidate for use as a subunit vaccine. Specifically, αvβ6- and αvβ8-integrins serve as receptors for HSV entry into keratinocytes and other epithelial and neuronal cells. Both bind gH-gL with high affinity.
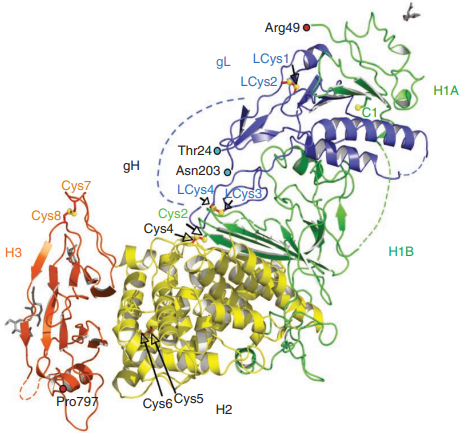
Fig. 5 Crystal structure of gH-gL complex(Chowdary et al., 2010)
Reference
1. AlMukdad, S., Harfouche, M., Wettstein, A., and Abu-Raddad, L.J. (2021). Epidemiology of herpes simplex virus type 2 in Asia: A systematic review, meta-analysis, and meta-regression. The Lancet Regional Health–Western Pacific 12.
2. Arduino, P.G., and Porter, S.R. (2008). Herpes Simplex Virus Type 1 infection: overview on relevant clinico-pathological features. J Oral Pathol Med 37, 107-121.
3. Bai, L., Xu, J., Zeng, L., Zhang, L., and Zhou, F. (2024). A review of HSV pathogenesis, vaccine development, and advanced applications. Molecular Biomedicine 5, 35.
4. Bernstein, D.I., Bellamy, A.R., Hook, E.W., III, Levin, M.J., Wald, A., Ewell, M.G., Wolff, P.A., Deal, C.D., Heineman, T.C., Dubin, G., et al. (2012). Epidemiology, Clinical Presentation, and Antibody Response to Primary Infection With Herpes Simplex Virus Type 1 and Type 2 in Young Women. Clinical Infectious Diseases 56, 344-351.
5. Chowdary, T.K., Cairns, T.M., Atanasiu, D., Cohen, G.H., Eisenberg, R.J., and Heldwein, E.E. (2010). Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol 17, 882-888.
6. Cooper, R.S., Georgieva, E.R., Borbat, P.P., Freed, J.H., and Heldwein, E.E. (2018). Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB. Nat Struct Mol Biol 25, 416-424.
7. Dai, X., and Zhou, Z.H. (2018). Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 360, eaao7298.
8. Di Giovine, P., Settembre, E.C., Bhargava, A.K., Luftig, M.A., Lou, H., Cohen, G.H., Eisenberg, R.J., Krummenacher, C., and Carfi, A. (2011). Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS pathogens 7, e1002277.
9. Hoog, S.S., Smith, W.W., Qiu, X., Janson, C.A., Hellmig, B., McQueney, M.S., O'Donnell, K., O'Shannessy, D., DiLella, A.G., Debouck, C., et al. (1997). Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex. Biochemistry 36, 14023-14029.
10. Jama, M., Owen, E.M., Nahal, B., Obasi, A., and Clarke, E. (2024). Twenty years of herpes simplex virus type 2 (HSV-2) research in low-income and middle-income countries: systematic evaluation of progress made in addressing WHO prioritiesfor research in HSV-2 epidemiology and diagnostics. BMJ Global Health 9, e012717.
11. Jambunathan, N., Clark, C.M., Musarrat, F., Chouljenko, V.N., Rudd, J., and Kousoulas, K.G. (2021). Two Sides to Every Story: Herpes Simplex Type-1 Viral Glycoproteins gB, gD, gH/gL, gK, and Cellular Receptors Function as Key Players in Membrane Fusion. Viruses 13, 1849.
12. James, C., Harfouche, M., Welton, N.J., Turner, K.M., Abu-Raddad, L.J., Gottlieb, S.L., and Looker, K.J. (2020). Herpes simplex virus: global infection prevalence and incidence estimates, 2016. Bulletin of the World Health Organization 98, 315.
13. Liu, Y.T., Jih, J., Dai, X., Bi, G.Q., and Zhou, Z.H. (2019). Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome. Nature 570, 257-261.
14. Looker, K.J., and Garnett, G.P. (2005). A systematic review of the epidemiology and interaction of herpes simplex virus types 1 and 2. Sex Transm Infect 81, 103-107.
15. Peng, T., Ponce-de-Leon, M., Jiang, H., Dubin, G., Lubinski, J.M., Eisenberg, R.J., and Cohen, G.H. (1998). The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J Virol 72, 65-72.
16. Sari, T.K., Gianopulos, K.A., and Nicola, A.V. (2020). Glycoprotein C of Herpes Simplex Virus 1 Shields Glycoprotein B from Antibody Neutralization. Journal of Virology 94, 10.1128/jvi.01852-01819.
17. Sprague, E.R., Wang, C., Baker, D., and Bjorkman, P.J. (2006). Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging. PLoS Biol 4, e148.
18. Stampfer, S.D., Lou, H., Cohen, G.H., Eisenberg, R.J., and Heldwein, E.E. (2010). Structural Basis of Local, pH-Dependent Conformational Changes in Glycoprotein B from Herpes Simplex Virus Type 1. Journal of Virology 84, 12924-12933.
19. Zhang, N., Yan, J., Lu, G., Guo, Z., Fan, Z., Wang, J., Shi, Y., Qi, J., and Gao, G.F. (2011). Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion. Nat Commun 2, 577.
20. Zhu, S., and Viejo-Borbolla, A. (2021). Pathogenesis and virulence of herpes simplex virus. Virulence 12, 2670-2702.
Sample type: Plasma, Serum
Host species: Alpaca (Lama pacos)
Isotype: VHH-8His-Cys-tag
Applications: ELISA
Accession: A1Z0P7, P06763
Applications: Used for the quantitative determination of HSV-1 gD concentration in serum and plasma.
Sample type: Plasma, Serum
Sensitivity: 0.08 ng/mL
Range: 0.78 - 50 ng/mL
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: P10205
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: P10202
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: P32888