(Sharma, Suresh. (2010). Hepatitis C virus: Molecular biology & current therapeutic options. The Indian journal of medical research. 131. 17-34.)
Hepatitis C Virus
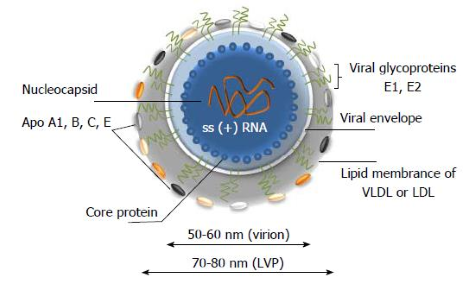
Hepatitis C virus (HCV) is an enveloped, positive-strand RNA virus classified in the Hepacivirus genus within the Flaviviridae family.
HCV genomic RNA is packaged by core protein and enveloped by a lipid bilayer containing two viral glycoproteins (E1 and E2) to form the virion.
The HCV lifecycle begins with the attachment of a virion to its specific receptors on hepatocytes. Up to now, low density lipoprotein receptor (LDLr) and heparin sulfate proteoglycans (HSPGs), scavenger receptor class B type I (SRB1), tetraspanin CD81, tight junction protein claudin-1(CLDN1) are the known cellular receptors initiating the attachment step of HCV infection. It is proposed that the virus, after binding with its receptor complex, is internalized, and that the nucleocapsid is released into the cytoplasm. The virus is then uncoating to free its genomic RNA, and the HCV genomic RNA is used both for polyprotein translation and replication in the cytoplasm.
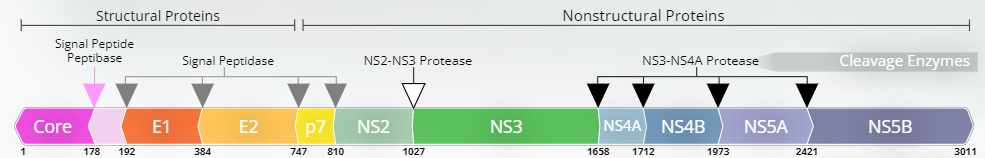
The translation process generates a single polyprotein that is approximately 3,000 amino acids long, which will be further processed by host and viral proteases, resulting in 10 different HCV proteins (core, E1, E2, p7, NS3, NS4A, NS4B, NS5A, NS5B).
Genome polyprotein (P26664)
Function:
The genome polyprotein is approximately 3,000 amino acids long. It will be further processed by host and viral proteases to produce 10 mature HCV proteins (core, E1, E2, p7, NS3, NS4A, NS4B, NS5A, NS5B).
Packages viral RNA to form a viral nucleocapsid, and promotes virion budding (Probable). Participates in the viral particle production as a result of its interaction with the non-structural protein 5A (By similarity).Binds RNA and may function as a RNA chaperone to induce the RNA structural rearrangements taking place during virus replication (By similarity).Modulates viral translation initiation by interacting with viral IRES and 40S ribosomal subunit (By similarity).
Affects various cell signaling pathways, host immunity and lipid metabolism (Probable). Prevents the establishment of cellular antiviral state by blocking the interferon-alpha/beta (IFN-alpha/beta) and IFN-gamma signaling pathways and by blocking the formation of phosphorylated STAT1 and promoting ubiquitin-mediated proteasome-dependent degradation of STAT1 (By similarity).
Forms a heterodimer with envelope glycoprotein E2, which mediates virus attachment to the host cell, virion internalization through clathrin-dependent endocytosis and fusion with host membrane (By similarity).
Fusion with the host cell is most likely mediated by both E1 and E2, through conformational rearrangements of the heterodimer required for fusion rather than a classical class II fusion mechanism (By similarity).
E1/E2 heterodimer binds host apolipoproteins such as APOB and APOE thereby forming a lipo-viro-particle (LVP) (By similarity).
APOE associated to the LVP allows the initial virus attachment to cell surface receptors such as the heparan sulfate proteoglycans (HSPGs), syndecan-1 (SDC1), syndecan-1 (SDC2), the low-density lipoprotein receptor (LDLR) and scavenger receptor class B type I (SCARB1) (By similarity).
The cholesterol transfer activity of SCARB1 allows E2 exposure and binding of E2 to SCARB1 and the tetraspanin CD81 (By similarity).
E1/E2 heterodimer binding on CD81 activates the epithelial growth factor receptor (EGFR) signaling pathway (By similarity).
Diffusion of the complex E1-E2-EGFR-SCARB1-CD81 to the cell lateral membrane allows further interaction with Claudin 1 (CLDN1) and occludin (OCLN) to finally trigger HCV entry (By similarity).
Forms a heterodimer with envelope glycoprotein E1, which mediates virus attachment to the host cell, virion internalization through clathrin-dependent endocytosis and fusion with host membrane (By similarity).
Fusion with the host cell is most likely mediated by both E1 and E2, through conformational rearrangements of the heterodimer required for fusion rather than a classical class II fusion mechanism (By similarity).
The interaction between envelope glycoprotein E2 and host apolipoprotein E/APOE allows the proper assembly, maturation and infectivity of the viral particles (By similarity).
This interaction is probably promoted via the up-regulation of cellular autophagy by the virus (By similarity).
E1/E2 heterodimer binds host apolipoproteins such as APOB and APOE thereby forming a lipo-viro-particle (LVP) (By similarity).
APOE associated to the LVP allows the initial virus attachment to cell surface receptors such as the heparan sulfate proteoglycans (HSPGs), syndecan-1 (SDC1), syndecan-1 (SDC2), the low-density lipoprotein receptor (LDLR) and scavenger receptor class B type I (SCARB1) (By similarity).
The cholesterol transfer activity of SCARB1 allows E2 exposure and binding of E2 to SCARB1 and the tetraspanin CD81 (By similarity).
E1/E2 heterodimer binding on CD81 activates the epithelial growth factor receptor (EGFR) signaling pathway (By similarity).
Diffusion of the complex E1-E2-EGFR-SCARB1-CD81 to the cell lateral membrane allows further interaction with Claudin 1 (CLDN1) and occludin (OCLN) to finally trigger HCV entry (By similarity).
Inhibits host EIF2AK2/PKR activation, preventing the establishment of an antiviral state (PubMed:10390359, PubMed:11152499).
Viral ligand for CD209/DC-SIGN and CLEC4M/DC-SIGNR, which are respectively found on dendritic cells (DCs), and on liver sinusoidal endothelial cells and macrophage-like cells of lymph node sinuses (By similarity).
These interactions allow the capture of circulating HCV particles by these cells and subsequent facilitated transmission to permissive cells such as hepatocytes and lymphocyte subpopulations (By similarity).
The interaction between E2 and host amino acid transporter complex formed by SLC3A2 and SLC7A5/LAT1 may facilitate viral entry into host cell (By similarity).
Ion channel protein that acts as a viroporin and plays an essential role in the assembly, envelopment and secretion of viral particles (By similarity).
Regulates the host cell secretory pathway, which induces the intracellular retention of viral glycoproteins and favors assembly of viral particles (By similarity).Creates a pore in acidic organelles and releases Ca2+ and H+ in the cytoplasm of infected cells, leading to a productive viral infection (By similarity).High levels of cytoplasmic Ca2+ may trigger membrane trafficking and transport of viral ER-associated proteins to viroplasms, sites of viral genome replication (Probable). This ionic imbalance induces the assembly of the inflammasome complex, which triggers the maturation of pro-IL-1beta into IL-1beta through the action of caspase-1 (By similarity).
Targets also host mitochondria and induces mitochondrial depolarization (By similarity).
In addition of its role as a viroporin, acts as a lipid raft adhesion factor (By similarity).
Cysteine protease required for the proteolytic auto-cleavage between the non-structural proteins NS2 and NS3 (By similarity).
The N-terminus of NS3 is required for the function of NS2 protease (active region NS2-3) (By similarity).
Promotes the initiation of viral particle assembly by mediating the interaction between structural and non-structural proteins (By similarity).
Displays three enzymatic activities: serine protease with a chymotrypsin-like fold, NTPase and RNA helicase (By similarity).
NS3 serine protease, in association with NS4A, is responsible for the cleavages of NS3-NS4A, NS4A-NS4B, NS4B-NS5A and NS5A-NS5B (By similarity).
The NS3/NS4A complex prevents phosphorylation of host IRF3, thus preventing the establishment of dsRNA induced antiviral state (By similarity).
The NS3/NS4A complex induces host amino acid transporter component SLC3A2, thus contributing to HCV propagation (By similarity).
NS3 RNA helicase binds to RNA and unwinds both dsDNA and dsRNA in the 3' to 5' direction, and likely resolves RNA complicated stable secondary structures in the template strand (By similarity).
Binds a single ATP and catalyzes the unzipping of a single base pair of dsRNA (By similarity).
Inhibits host antiviral proteins TBK1 and IRF3 thereby preventing the establishment of an antiviral state (By similarity).
Cleaves host MAVS/CARDIF thereby preventing the establishment of an antiviral state (By similarity).
Cleaves host TICAM1/TRIF, thereby disrupting TLR3 signaling and preventing the establishment of an antiviral state (By similarity).
The NS3/NS4A complex prevents phosphorylation of host IRF3, thus preventing the establishment of dsRNA induced antiviral state (By similarity).The NS3/NS4A complex induces host amino acid transporter component SLC3A2, thus contributing to HCV propagation (By similarity).
Induces a specific membrane alteration that serves as a scaffold for the virus replication complex (By similarity).This membrane alteration gives rise to the so-called ER-derived membranous web that contains the replication complex (By similarity).NS4B self-interaction contributes to its function in membranous web formation (By similarity).Promotes host TRIF protein degradation in a CASP8-dependent manner thereby inhibiting host TLR3-mediated interferon signaling (By similarity).Disrupts the interaction between STING and TBK1 contributing to the inhibition of interferon signaling (By similarity).
Phosphorylated protein that is indispensable for viral replication and assembly (By similarity).Both hypo- and hyperphosphorylated states are required for the viral life cycle (By similarity).The hyperphosphorylated form of NS5A is an inhibitor of viral replication (By similarity).Involved in RNA-binding and especially in binding to the viral genome (By similarity).Zinc is essential for RNA-binding (By similarity).Participates in the viral particle production as a result of its interaction with the mature viral core protein (By similarity).Its interaction with host VAPB may target the viral replication complex to vesicles (By similarity).
Down-regulates viral IRES translation initiation (PubMed:15784895).
Mediates interferon resistance, presumably by interacting with and inhibiting host EIF2AK2/PKR (PubMed:9143277).Prevents BIN1-induced apoptosis (By similarity).
Acts as a transcriptional activator of some host genes important for viral replication when localized in the nucleus (By similarity).
Via the interaction with host PACSIN2, modulates lipid droplet formation in order to promote virion assembly (By similarity).
Modulates TNFRSF21/DR6 signaling pathway for viral propagation (By similarity).
Nonstructural protein 5B (NS5B) is an RNA-dependent RNA polymerase (RdRp) that performs primer-template recognition and RNA synthesis during viral replication.
Reference:
Host species: Human
Isotype: IgG1
Applications: ELISA, Neutralization
Accession: P27958
Host species: Human
Isotype: IgG1
Applications: ELISA, Neutralization
Accession: P27958
Host species: Human
Isotype: IgG1
Applications: ELISA, Neutralization
Accession: P27958
Host species: Human
Isotype: IgG1
Applications: ELISA, Neutralization
Accession: P27958
Host species: Human
Isotype: IgG1
Applications: ELISA, Neutralization
Accession: P27958
Host species: Human
Isotype: IgG1
Applications: ELISA, Neutralization
Accession: P27958
Host species: Human
Isotype: IgG1
Applications: ELISA, Neutralization
Accession: P27958
Host species: Human
Isotype: IgG1
Applications: ELISA, Neutralization
Accession: P27958
Applications: ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Expression system: E. coli
Accession: A0A0S2CFQ4
Protein length: Phe10-Val172
Applications: ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Expression system: E. coli
Accession: P27958
Protein length: Glu384-Glu661