Catalog No.
YXX04901
Description
Peptide -N-Glycosidase F, also known as PNGase F, is an amidase that cleaves between the innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex oligosaccharides from N-linked glycoproteins
Biological activity
One unit is defined as the amount of enzyme required to remove > 95% of the carbohydrate from 10 μg of denatured RNase B in 1 hour at 37°C in a total reaction volume of 10 μl.
Expression system
E. coli
Species
Elizabethkingia miricola
Protein length
PNGase F is cloned from Elizabethkingia miricola and expressed in E.coli.
Predicted molecular weight
37.08 kDa
Nature
Recombinant
Target
PNGase F
Concentration
50,000 units/ml
Endotoxin level
Please contact with the lab for this information.
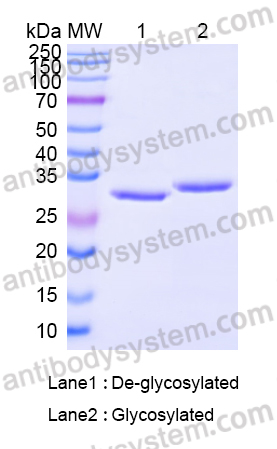
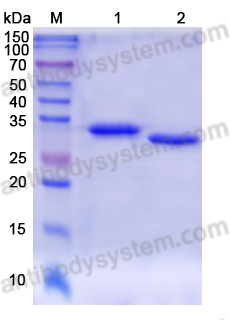
Purity
>95% as determined by SDS-PAGE.
Applications
Removal of high mannose N-glycans from glycoproteins
Form
Liquid
Storage buffer
20mM Tris-HCl, pH 7.5, 50mM NaCl, 5mM Na2EDTA, 50% glycerol
Experimental Procedure
Standard Operating Procedure
Denaturing Reaction Conditions:
1. Combine 1-20 µg of glycoprotein, 1 µl of Glycoprotein Denaturing Buffer (10X) and H2O (if necessary) to make a 10 µl total reaction volume.
2. Denature glycoprotein by heating reaction at 100°C for 10 minutes.
3. Chill denatured glycoprotein on ice and centrifuge 10 seconds.
4. Make a total reaction volume of 20 µl by adding 2 µl GlycoBuffer 2 (10X), 2 µl 10% NP-40 and 6 µl H2O.
5. Add 1 µl PNGase F, mix gently.
6. Incubate reaction at 37°C for 1 hour.
Note: The simplest method of assessing the extent of deglycosylation is by mobility shifts on SDS-PAGE gels.
Note: To deglycosylate different glycoprotein, longer incubation time may be required.
Non-Denaturing Reaction Conditions:
1.Combine 1-20 µg of glycoprotein, 2 µl of GlycoBuffer 2 (10X) and H2O (if necessary) to make a 20 µl total reaction volume.
2.Add 2-5 µl PNGase F, mix gently.
3.Incubate reaction at 37°C for 4 - 24 hours.
Note: To deglycosylate a native glycoprotein, longer incubation time as well as more enzyme may be required.
Note: The simplest method of assessing the extent of deglycosylation is by mobility shifts on SDS-PAGE gels.
1X Glycoprotein Denaturing Buffer
0.5% SDS
40 mM DTT
1X NP-40
1% NP-40 in MilliQ-H2O
1X GlycoBuffer 2
20 mM Tris,PH 7.5
Shipping
In general, proteins are provided as lyophilized powder/frozen liquid. They are shipped out with dry ice/blue ice unless customers require otherwise.
Stability and Storage
Use a manual defrost freezer and avoid repeated freeze thaw cycles.Store at 2 to 8 °C for one week .Store at -20 to -80 °C for twelve months from the date of receipt.
N-Glycosylation as a Key Requirement for the Positive Interaction of Integrin and uPAR in Glioblastoma., PMID:40508119
Impaired Proteostasis is Linked to Neurological Pathology in a Zebrafish NGLY1 Deficiency Model., PMID:40470739
Comparative Analysis of Serum N-Glycosylation in Endometriosis and Gynecologic Cancers., PMID:40362345
Aberrant glycosylation of IgG in children with active lupus nephritis alters podocyte metabolism and causes podocyte injury., PMID:40297959
Increased oxidative stress and autophagy in NGLY1 patient iPSC-derived neural stem cells., PMID:40189184
Development and characterisation of a method for determining N-terminal heterogeneity in the human recombinant follitropin β-subunit., PMID:40139043
PNGase activity and free N-glycans in phloem fluid prepared from Nerium oleander (oleander tree)., PMID:40132999
Enhanced Production and Functional Characterization of Recombinant Equine Chorionic Gonadotropin (rec-eCG) in CHO-DG44 Cells., PMID:40001592
Nucleolin in the cell membrane promotes Ang II-mediated VSMC phenotypic switching by regulating the AT1R internalization function : Nucleolin promotes Ang II-mediated VSMC phenotypic switching., PMID:40001211
Biochemical characterization of the Escherichia coli surfaceome: a focus on type I fimbriae and flagella., PMID:39973929
PNGaseF-Generated N-Glycans Adduct onto Peptides in the Gas Phase., PMID:39938005
Altered Pattern of Serum N-Glycome in Subarachnoid Hemorrhage and Cerebral Vasospasm., PMID:39860471
The Glycosylation of Serum IgG Antibodies in Post-COVID-19 and Post-Vaccination Patients., PMID:39859521
Expression and purification of PNGase F protein in yeast and its anti-PRV activity., PMID:39827598
Preparative Isolation of N-Glycans from Natural Sources Mediated by a Deglycosylating Heterogeneous Biocatalyst in Flow., PMID:39817794
Decellularization and Enzymatic Digestion Methods to Enhance ECM Protein Detection via MALDI-MS Imaging., PMID:39753373
Hypoxia-induced TPC2 transcription and glycosylation aggravates pulmonary arterial hypertension by blocking autophagy flux., PMID:39732974
Correction to "The kinase domains of obscurin interact with intercellular adhesion proteins"., PMID:39651913
Glycosylation analysis of transcription factor TFIIB using bioinformatics and experimental methods., PMID:39601751
Site-Specific N-Glycoprofiling of Immunoglobulin G Subtypes from Donkey Milk., PMID:39557633
Characterization of Cell Surface Glycoproteins Using Enzymatic Treatment and Mass Spectrometry., PMID:39556700
3D structural insights into the effect of N-glycosylation in human chitotriosidase variant G102S., PMID:39521151
Expression and immobilization of novel N-glycan-binding protein for highly efficient purification and enrichment of N-glycans, N-glycopeptides, and N-glycoproteins., PMID:39412696
Oxidative Release of Natural Glycans: Unraveling the Mechanism for Rapid N-Glycan Glycomics Analysis., PMID:39387489
Identification of N-linked glycans recognized by WGA in saliva from patients with non-small cell lung cancer., PMID:39241666
Development of new NGLY1 assay systems - toward developing an early screening method for NGLY1 deficiency., PMID:39206713
Structural basis of sugar recognition by SCFFBS2 ubiquitin ligase involved in NGLY1 deficiency., PMID:39171510
Systemic gene therapy corrects the neurological phenotype in a mouse model of NGLY1 deficiency., PMID:39137042
A proton channel, Otopetrin 1 (OTOP1) is N-glycosylated at two asparagine residues in third extracellular loop., PMID:39129225
A chemoenzymatic method for simultaneous profiling N- and O-glycans on glycoproteins using one-pot format., PMID:39116882
The SCR-17 and SCR-18 glycans in human complement factor H enhance its regulatory function., PMID:39098532
Direct derivatization of sialic acids and mild β-elimination for linkage-specific sialyl O-glycan analysis., PMID:39067924
N-Glycosylation of MRS2 balances aerobic and anaerobic energy production by reducing rapid mitochondrial Mg2+ influx in conditions of high glucose or impaired respiratory chain function., PMID:39026824
Stable Production of a Recombinant Single-Chain Eel Follicle-Stimulating Hormone Analog in CHO DG44 Cells., PMID:39000389
Mutations in nucleotide metabolism genes bypass proteasome defects in png-1/NGLY1-deficient Caenorhabditis elegans., PMID:38991033
Rapid Mass Spectrometry-Based Multiattribute Method for Glycation Analysis with Integrated Afucosylation Detection Capability., PMID:38970800
PD-L1 has a heterogeneous and dynamic expression in gastric cancer with implications for immunoPET., PMID:38915392
Characterizing Glycosylation of Adeno-Associated Virus Serotype 9 Capsid Proteins Generated from HEK293 Cells through Glycopeptide Mapping and Released Glycan Analysis., PMID:38792776
Doxorubicin induces deglycosylation of cancer cell-intrinsic PD-1 by NGLY1., PMID:38782868
Surface plasmon resonance microscopy identifies glycan heterogeneity in pancreatic cancer cells that influences mucin-4 binding interactions., PMID:38776309
Ocular features of NGLY1 deficiency from a prospective longitudinal cohort., PMID:38697387
Intranasal oxytocin suppresses seizure-like behaviors in a mouse model of NGLY1 deficiency., PMID:38649481
ELISA-based highly sensitive assay system for the detection of endogenous NGLY1 activity., PMID:38581946
Mass spectrometry imaging protocol for spatial mapping of lipids, N-glycans and peptides in murine lung tissue., PMID:38525810
Site-Specific Conjugation of Native Antibody: Transglutaminase-Mediated Modification of a Conserved Glutamine While Maintaining the Primary Sequence and Core Fc Glycan via Trimming with an Endoglycosidase., PMID:38499390
Updates of the current strategies of labeling for N-glycan analysis., PMID:38484674
Complex N-glycosylation of mGluR6 is required for trans-synaptic interaction with ELFN adhesion proteins., PMID:38428819
CSF N-Glycomics Using MALDI MS Techniques., PMID:38427187
Development of a fluorescence and quencher-based FRET assay for detection of endogenous peptide:N-glycanase/NGLY1 activity., PMID:38417795
A commentary on 'Patient-derived gene and protein expression signatures of NGLY1 deficiency'., PMID:38156787