Catalog No.
YVV01501
Expression system
E. coli
Species
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
Protein length
Asn4141-Gln4253
Predicted molecular weight
14.67 kDa
Nature
Recombinant
Endotoxin level
Please contact with the lab for this information.
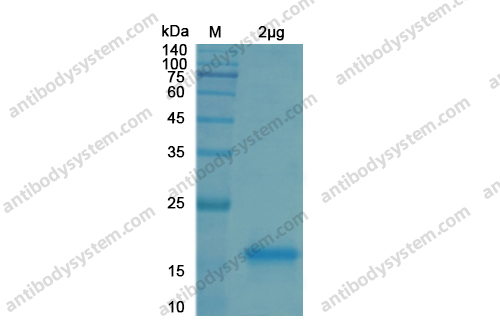
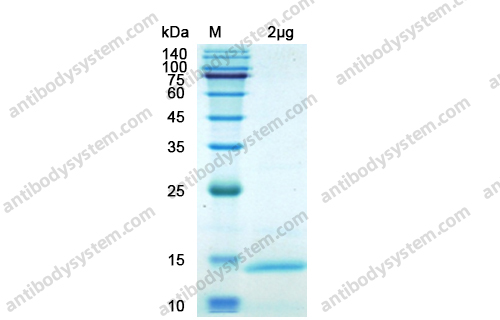
Purity
>90% as determined by SDS-PAGE.
Accession
P0DTD1
Applications
ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Form
Lyophilized
Storage buffer
Lyophilized from a solution in PBS pH 7.4, 0.02% NLS, 1mM EDTA, 4% Trehalose, 1% Mannitol.
Reconstitution
Reconstitute in sterile water for a stock solution. A copy of datasheet will be provided with the products, please refer to it for details.
Shipping
In general, proteins are provided as lyophilized powder/frozen liquid. They are shipped out with dry ice/blue ice unless customers require otherwise.
Stability and Storage
Use a manual defrost freezer and avoid repeated freeze thaw cycles. Store at 2 to 8°C for frequent use. Store at -20 to -80°C for twelve months from the date of receipt.
Alternative Names
Non-structural protein 9, nsp9, Replicase polyprotein 1ab, pp1ab, ORF1ab polyprotein
Defining short linear motif binding determinants by phage display-based deep mutational scanning., PMID:40411416
Virtual Screening of Phytoconstituents in Indian Spices Based on their Inhibitory Potential against SARS-CoV-2., PMID:40353411
Peptide nucleic acids can form hairpins and bind RNA-binding proteins., PMID:39283902
Nanobody against SARS-CoV-2 non-structural protein Nsp9 inhibits viral replication in human airway epithelia., PMID:39281707
Mapping the immunopeptidome of seven SARS-CoV-2 antigens across common HLA haplotypes., PMID:39214998
Comparative Atlas of SARS-CoV-2 Substitution Mutations: A Focus on Iranian Strains Amidst Global Trends., PMID:39205305
One pot multi-component synthesis of novel functionalized pyrazolo furan-2(5H)-one derivatives: in vitro, DFT, molecular docking, and pharmacophore studies, as coronavirus inhibitors., PMID:39168959
A Biochemical and Biophysical Analysis of the Interaction of nsp9 with nsp12 from SARS-CoV-2-Implications for Future Drug Discovery Efforts., PMID:38958516
Investigating SARS-CoV-2 virus-host interactions and mRNA expression: Insights using three models of D. melanogaster., PMID:38925484
SARS-CoV-2 Protein Nsp9 Is Involved in Viral Evasion through Interactions with Innate Immune Pathways., PMID:38911767
Inhibition of SARS-CoV-2 Nsp9 ssDNA-Binding Activity and Cytotoxic Effects on H838, H1975, and A549 Human Non-Small Cell Lung Cancer Cells: Exploring the Potential of Nepenthes miranda Leaf Extract for Pulmonary Disease Treatment., PMID:38892307
Mutations in nonstructural proteins essential for pathogenicity in SARS-CoV-2-infected mice., PMID:38888344
Mutation prediction in the SARS-CoV-2 genome using attention-based neural machine translation., PMID:38872567
Genomic Context of SARS-CoV-2 Outbreaks in Farmed Mink in Spain during Pandemic: Unveiling Host Adaptation Mechanisms., PMID:38791536
Assessing nanobody interaction with SARS-CoV-2 Nsp9., PMID:38758765
Adaptation of SARS-CoV-2 to ACE2H353K mice reveals new spike residues that drive mouse infection., PMID:38168680
Discovery of Anti-SARS-CoV-2 Nsp9 Binders from Natural Products by a Native Mass Spectrometry Approach., PMID:37993134
A structural analysis of the nsp9 protein from the coronavirus MERS CoV reveals a conserved RNA binding interface., PMID:37929701
SARS-COV-2 protein NSP9 promotes cytokine production by targeting TBK1., PMID:37854611
SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9., PMID:37794589
Identification of motif-based interactions between SARS-CoV-2 protein domains and human peptide ligands pinpoint antiviral targets., PMID:37704626
Conserved Characteristics of NMPylation Activities of Alpha- and Betacoronavirus NiRAN Domains., PMID:37199624
Collision-Induced Affinity Selection Mass Spectrometry for Identification of Ligands., PMID:37101899
SARS-CoV-2 polyprotein substrate regulates the stepwise Mpro cleavage reaction., PMID:37044215
Inside-out: Antibody-binding reveals potential folding hinge-points within the SARS-CoV-2 replication co-factor nsp9., PMID:37036856
Global landscape of SARS-CoV-2 mutations and conserved regions., PMID:36841805
Genomic landscape of alpha-variant of SARS-CoV-2 circulated in Pakistan., PMID:36512569
Virtual screening of phytochemicals by targeting multiple proteins of severe acute respiratory syndrome coronavirus 2: Molecular docking and molecular dynamics simulation studies., PMID:36442514
Determination of binding affinity of tunicamycin with SARS-CoV-2 proteins: Proteinase, protease, nsp2, nsp9, ORF3a, ORF7a, ORF8, ORF9b, envelope and RBD of spike glycoprotein., PMID:36349218
A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors., PMID:36335936
SARS-CoV-2 mitochondriopathy in COVID-19 pneumonia exacerbates hypoxemia., PMID:36334378
Structural modeling of protein ensembles between E3 RING ligases and SARS-CoV-2: The role of zinc binding domains., PMID:36209710
The mechanism of RNA capping by SARS-CoV-2., PMID:35944563
Affinity selection-mass spectrometry in the discovery of anti-SARS-CoV-2 compounds., PMID:35929396
Expression Profile and Localization of SARS-CoV-2 Nonstructural Replicase Proteins in Infected Cells., PMID:35730969
SARS-CoV-2 couples evasion of inflammatory response to activated nucleotide synthesis., PMID:35700355
Discovery and functional interrogation of SARS-CoV-2 protein-RNA interactions., PMID:35313591
Discovery and functional interrogation of SARS-CoV-2 protein-RNA interactions., PMID:35233578
Collection of Monoclonal Antibodies Targeting SARS-CoV-2 Proteins., PMID:35216036
The mechanism of RNA capping by SARS-CoV-2., PMID:35194601
Shedding Light on the Synthesis, Crystal Structure, Characterization, and Computational Study of Optoelectronic Properties and Bioactivity of Imine derivatives., PMID:35187337
Anti-SARS-CoV-2 IgG responses are powerful predicting signatures for the outcome of COVID-19 patients., PMID:35116173
NSP9 of SARS-CoV-2 attenuates nuclear transport by hampering nucleoporin 62 dynamics and functions in host cells., PMID:34844119
Structural, energetic and lipophilic analysis of SARS-CoV-2 non-structural protein 9 (NSP9)., PMID:34837010
A natural product compound inhibits coronaviral replication in vitro by binding to the conserved Nsp9 SARS-CoV-2 protein., PMID:34756886
NMR-Based Analysis of Nanobodies to SARS-CoV-2 Nsp9 Reveals a Possible Antiviral Strategy Against COVID-19., PMID:34705339
Polypharmacology of some medicinal plant metabolites against SARS-CoV-2 and host targets: Molecular dynamics evaluation of NSP9 RNA binding protein., PMID:34370622
A distinct ssDNA/RNA binding interface in the Nsp9 protein from SARS-CoV-2., PMID:34369011
NMPylation and de-NMPylation of SARS-CoV-2 nsp9 by the NiRAN domain., PMID:34352100
Binding of a pyrimidine RNA base-mimic to SARS-CoV-2 nonstructural protein 9., PMID:34331944