Catalog No.
EVV00201
Expression system
Mammalian Cells
Species
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
Protein length
Ser13-Leu303
Predicted molecular weight
36.23KD
Nature
Recombinant
Endotoxin level
Please contact with the lab for this information.
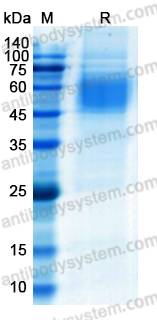
Purity
>85% as determined by SDS-PAGE.
Accession
P0DTC2
Applications
ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Form
Lyophilized
Storage buffer
Lyophilized from a solution in PBS pH 7.4, 1mM EDTA, 4% Trehalose, 1% Mannitol.
Reconstitution
Reconstitute in sterile water for a stock solution.A copy of datasheet will be provided with the products, please refer to it for details.
Shipping
In general, proteins are provided as lyophilized powder/frozen liquid. They are shipped out with dry ice/blue ice unless customers require otherwise.
Stability and Storage
Use a manual defrost freezer and avoid repeated freeze thaw cycles. Store at 2 to 8°C for frequent use. Store at -20 to -80°C for twelve months from the date of receipt.
Alternative Names
N-terminal domain, NTD, Spike glycoprotein, S glycoprotein, E2, Peplomer protein, Spike protein S1, S
Structural Analysis of the SARS-CoV-2 Spike N-Terminal Domain Across Wild-Type and Recent Variants: A Comparative Study., PMID:40485545
Is SARS-CoV-2 Spike Evolution Being Retargeted at the N-Terminal Domain?, PMID:40415355
Genome data artifacts and functional studies of deletion repair in the BA.1 SARS-CoV-2 spike protein., PMID:40308784
Dual-Locking the SARS-CoV-2 Spike Trimer: An Amphipathic Molecular "Bolt" Stabilizes Conserved Druggable Interfaces for Coronavirus Inhibition., PMID:40285637
Characterization of Nanobody Binding to Distinct Regions of the SARS-CoV-2 Spike Protein by Flow Virometry., PMID:40285013
Spike Protein-Fibrinogen Interaction: A Novel Immune Evasion Strategy of SARS-CoV-2?, PMID:40242590
Anti-S2 antibodies responsible for the SARS-CoV-2 infection-induced serological cross-reactivity against MERS-CoV and MERS-related coronaviruses., PMID:40226608
Allosteric Control and Glycan Shielding Adaptations in the SARS-CoV-2 Spike from Early to Peak Virulence., PMID:40161746
Cleaved vs. Uncleaved: How Furin Cleavage Reshapes the Conformational Landscape of SARS-CoV-2 Spike., PMID:40161653
Within-Host Fitness and Antigenicity Shift Are Key Factors Influencing the Prevalence of Within-Host Variations in the SARS-CoV-2 S Gene., PMID:40143291
Conformational and Stability Analysis of SARS-CoV-2 Spike Protein Variants by Molecular Simulation., PMID:40137759
Neutralization and spike stability of JN.1-derived LB.1, KP.2.3, KP.3, and KP.3.1.1 subvariants., PMID:40136024
Role of glycosylation mutations at the N-terminal domain of SARS-CoV-2 XEC variant in immune evasion, cell-cell fusion, and spike stability., PMID:40135879
SARS CoV-2 spike adopts distinct conformational ensembles in situ., PMID:40093071
Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern., PMID:40043139
Exploring the effects of N234 and N343 linked glycans to SARS CoV 2 spike protein pocket accessibility using Gaussian accelerated molecular dynamics simulations., PMID:40016249
NIEAs elicited by wild-type SARS-CoV-2 primary infection fail to enhance the infectivity of Omicron variants., PMID:39994733
Decoding omicron: Genetic insight into its transmission dynamics, severity spectrum and ever-evolving strategies of immune escape in comparison with other SARS-CoV-2 variants., PMID:39889436
Structural insights into nucleocapsid protein variability: Implications for PJ34 efficacy against SARS-CoV-2., PMID:39848104
Ancestral SARS-CoV-2 immune imprinting persists on RBD but not NTD after sequential Omicron infections., PMID:39807166
Genomic Evolution of the SARS-CoV-2 Omicron Variant in Córdoba, Argentina (2021-2022): Analysis of Uncommon and Prevalent Spike Mutations., PMID:39772187
Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants., PMID:39772146
Affinity Tag-Free Purification of SARS-CoV-2 N Protein and Its Crystal Structure in Complex with ssDNA., PMID:39766245
Development of a two-component recombinant vaccine for COVID-19., PMID:39759508
SARS-CoV-2 NSP3/4 control formation of replication organelle and recruitment of RNA polymerase NSP12., PMID:39737877
Enhanced antibody response to the conformational non-RBD region via DNA prime-protein boost elicits broad cross-neutralization against SARS-CoV-2 variants., PMID:39727342
Emergence of Omicron FN.1 a descendent of BQ.1.1 in Botswana., PMID:39720788
Structural serology of polyclonal antibody responses to mRNA-1273 and NVX-CoV2373 COVID-19 vaccines., PMID:39713412
Disruption of Molecular Interactions between the G3BP1 Stress Granule Host Protein and the Nucleocapsid (NTD-N) Protein Impedes SARS-CoV-2 Virus Replication., PMID:39708056
Cross-neutralization of distant coronaviruses correlates with Spike S2-specific antibodies from immunocompetent and immunocompromised vaccinated SARS-CoV-2-infected patients., PMID:39678346
Prompt engineering-enabled LLM or MLLM and instigative bioinformatics pave the way to identify and characterize the significant SARS-CoV-2 antibody escape mutations., PMID:39657873
A core network in the SARS-CoV-2 nucleocapsid NTD mediates structural integrity and selective RNA-binding., PMID:39653699
Protein nanoparticle vaccines induce potent neutralizing antibody responses against MERS-CoV., PMID:39644492
A potential bimodal interplay between heme and complement factor H 402H in the deregulation of the complement alternative pathway by SARS-CoV-2., PMID:39643072
Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain., PMID:39619228
Immune Evasion, Cell-Cell Fusion, and Spike Stability of the SARS-CoV-2 XEC Variant: Role of Glycosylation Mutations at the N-terminal Domain., PMID:39605558
Structural characterization of human monoclonal antibodies targeting uncommon antigenic sites on spike glycoprotein of SARS-CoV., PMID:39589795
Structure-function mapping and mechanistic insights on the SARS CoV2 Nsp1., PMID:39584680
Exploring B-cell epitope conservation and antigenicity shift in current COVID-19 variants: Analyzing spike-antibody interactions for therapeutic uses., PMID:39531907
SARS-CoV-2 nucleocapsid protein interaction with YBX1 displays oncolytic properties through PKM mRNA destabilization., PMID:39506849
Structural insight into broadening SARS-CoV-2 neutralization by an antibody cocktail harbouring both NTD and RBD potent antibodies., PMID:39470577
SARS-CoV-2 N protein coordinates viral particle assembly through multiple domains., PMID:39412257
Characterisation of the antibody-mediated selective pressure driving intra-host evolution of SARS-CoV-2 in prolonged infection., PMID:39405332
The HCoV-HKU1 N-Terminal Domain Binds a Wide Range of 9-O-Acetylated Sialic Acids Presented on Different Glycan Cores., PMID:39394950
A compact stem-loop DNA aptamer targets a uracil-binding pocket in the SARS-CoV-2 nucleocapsid RNA-binding domain., PMID:39380503
Omicron-specific ultra-potent SARS-CoV-2 neutralizing antibodies targeting the N1/N2 loop of Spike N-terminal domain., PMID:39361729
Expanding the tool box for native structural biology: 19F dynamic nuclear polarization with fast magic angle spinning., PMID:39356759
Isolation and characterization of IgG3 glycan-targeting antibodies with exceptional cross-reactivity for diverse viral families., PMID:39292703
Neutralization and Stability of JN.1-derived LB.1, KP.2.3, KP.3 and KP.3.1.1 Subvariants., PMID:39282390
N121T and N121S substitutions on the SARS-CoV-2 spike protein impact on serum neutralization., PMID:39221474