In molecular biology experiments, qPCR (quantitative real-time PCR) and Western blot (WB) are regarded as a complementary pair: qPCR detects transcriptional levels (mRNA), while WB validates protein expression. However, when results from these two methods contradict each other, researchers often encounter confusion—did the experiment fail, or is there an underlying biological phenomenon at play? Let us explore these mysteries with Dr. Connie.
Gene Expression ≠ Protein Expression: Temporal differences in gene expression
Principle: Gene expression is a dynamic process characterized by temporal delays between transcription (mRNA synthesis) and translation (protein synthesis). These delays can significantly impact the interpretation of experimental results. For instance:
Transcription precedes translation: Elevated mRNA levels detected by quantitative PCR (qPCR) may indicate early activation of a gene, peaking at 6 hours post-stimulation. However, the corresponding protein synthesis may require additional time, becoming significant only after 24 hours.
Delayed protein degradation: Some proteins, such as structural proteins, exhibit half-lives that span several days. Consequently, even if mRNA levels decrease (as indicated by negative qPCR results), residual protein can still be detected through Western blotting (WB).
Example: Following drug treatment, qPCR may reveal an mRNA peak at 6 hours, while WB analysis shows an increase in protein levels only after 24 hours.
Differences in experimental techniques
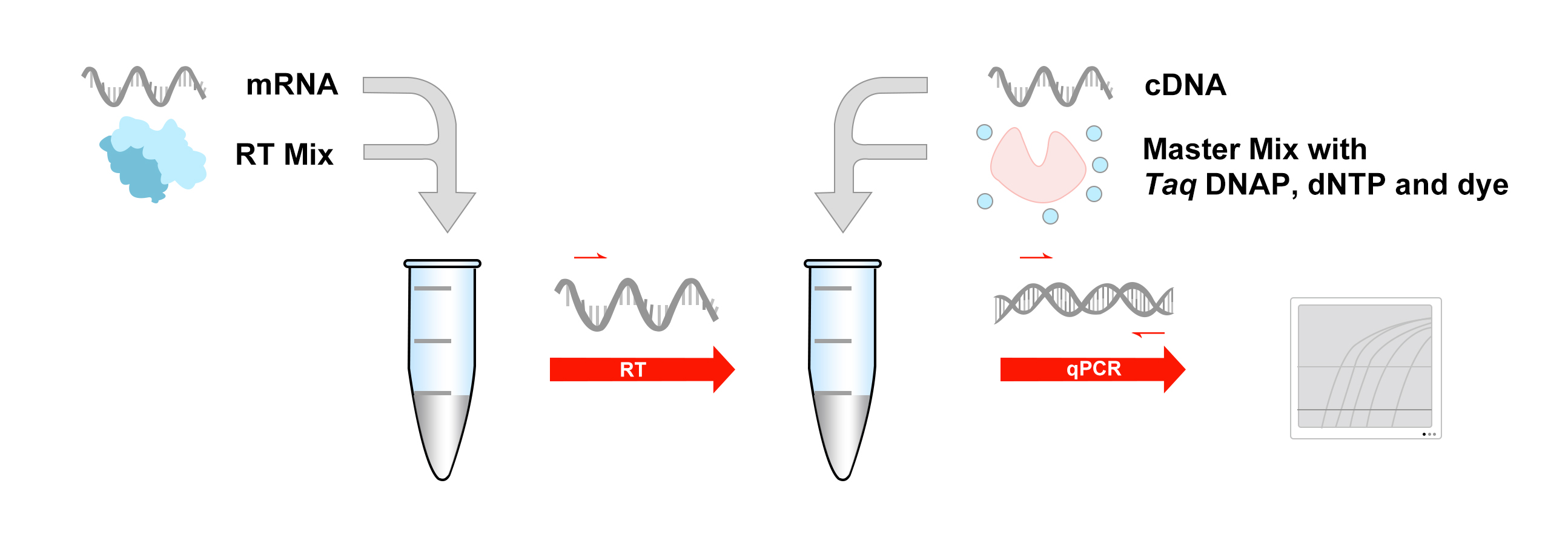
Sample handling: qPCR requires high-purity RNA (avoid RNase/DNA contamination), whereas protein extraction demands avoidance of denaturation or aggregation (e.g., membrane proteins require strong detergents).
Primer/antibody specificity issues: qPCR often uses housekeeping genes (e.g., GAPDH), while WB relies on β-actin. Fluctuations in reference gene expression can lead to normalization errors.
Tip: Always validate primer efficiency (via standard curve slopes) and antibody specificity (using knockout/knockdown models).
- Sample storage: Improper handling of aliquoted samples (e.g., repeated freeze-thaw cycles) may degrade RNA or denature proteins.
Example: A lab using WB on samples frozen-thawed three times at -80°C observed nonspecific bands, leading to false positives.
Sensitivity vs. specificity: qPCR detects mRNA at very low copy numbers but is sensitive to RNA quality (e.g., degradation). WB depends on antibody quality; low-abundance proteins or nonspecific binding may cause false negatives/positives.
Sample handling: qPCR requires high-purity RNA (avoid RNase/DNA contamination), whereas protein extraction demands avoidance of denaturation or aggregation (e.g., membrane proteins require strong detergents).
Primer/antibody specificity issues: qPCR often uses housekeeping genes (e.g., GAPDH), while WB relies on β-actin. Fluctuations in reference gene expression can lead to normalization errors.
The "Internal Reference Trap"
Poor internal reference selection: Using β-actin as a WB reference may fail if its expression changes under certain treatments (e.g., cytoskeletal reorganization).
qPCR "reference drift": Common reference genes (e.g., GAPDH, 18S rRNA) may be unstable under specific experimental conditions. Validate their stability beforehand.
Tip: Use Multiple internal reference (e.g., β-actin and GAPDH for WB) or tissue-specific references (e.g., TBP in cardiac tissue).
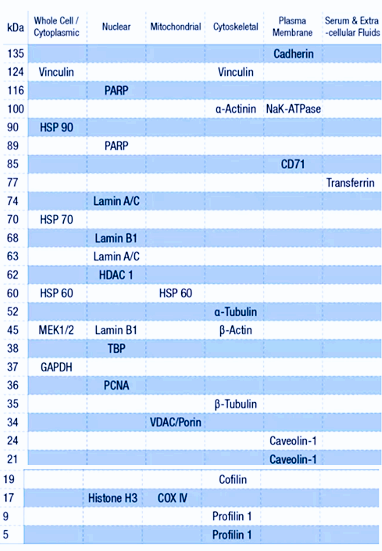
The internal references and their respective molecular weights associated with different cellular components
Protein Stability and Degradation
Principle: Protein half-lives vary significantly due to:
Ubiquitin-proteasome system: Short-lived proteins (e.g., p53, cyclins) degrade rapidly; even high mRNA levels may not yield detectable WB signals.
Lysosomal degradation: Membrane or misfolded proteins may be degraded via autophagy.
Post-translational modifications: Phosphorylation can tag proteins for degradation.
Example: Inflammatory cytokines (e.g., IL-6) exhibit transient mRNA spikes, but their proteins may be rapidly secreted or degraded, resulting in weak WB signals.
Translational Regulation
High mRNA levels do not guarantee increased protein levels. Translation can be regulated by:
Translational repression: mRNA may be bound and inhibited by miRNAs (common in cancer), preventing protein production.
Global translational suppression: Stress conditions (e.g., hypoxia, heat shock) trigger mechanisms like eIF2α phosphorylation to halt translation.
mRNA stability: Some mRNAs lack protective mechanisms (e.g., shortened poly-A tails) and degrade before translation.
Translational efficiency: Codon bias or RNA-binding proteins may alter protein output from the same mRNA.
Example: Oncogene mRNAs in tumor cells may fail to translate into proteins due to miRNA inhibition.
Post-Translational Modifications and Protein Activity
Principle: WB detects protein presence, not functional state:
Subcellular localization: Proteins may be secreted (e.g., cytokines) or localized to organelles (e.g., mitochondria). Incomplete lysis may prevent WB detection, causing mismatches with cytoplasmic mRNA levels.
Post-translational modifications: Phosphorylation or glycosylation may alter antibody epitopes (requiring modification-specific antibodies). Ubiquitination shifts molecular weights, potentially misclassifying bands as negative.
Example: The activation of signaling proteins (e.g., ERK) depends on phosphorylation; total ERK WB results may not correlate with qPCR mRNA levels.
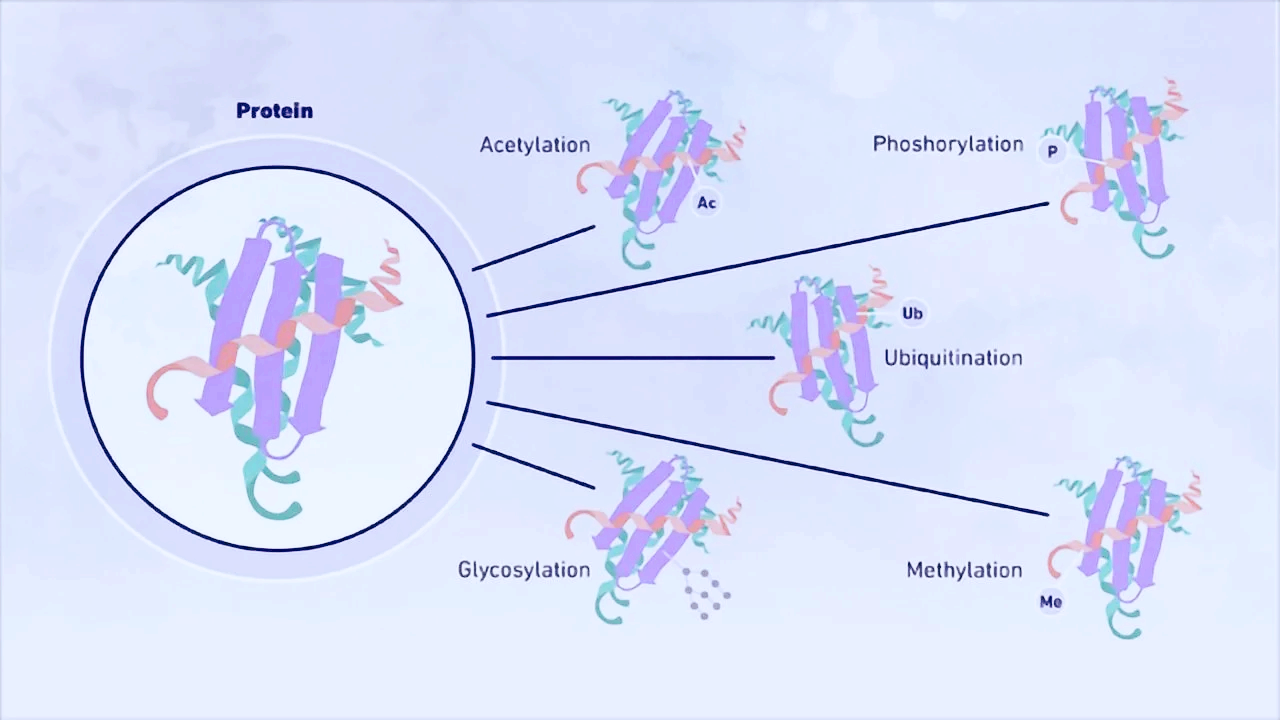
Post-translational modifications
Primer and Antibody Specificity Issues
Poor qPCR primer design (e.g., failure to span exon-exon junctions) may cause false negatives.
Cross-reactive WB antibodies (recognizing similar proteins) produce false-positive bands.
Tip: Validate primer efficiency and antibody specificity rigorously.
Table 1. Common Scenarios of Discordant Results
|
qPCR Result |
WB Result |
Potential Causes |
|
Increased |
Unchanged |
Translational repression, long protein half-life |
|
Unchanged |
Increased |
Enhanced translation, reduced protein degradation |
|
Increased |
Decreased |
Accelerated degradation (e.g., ubiquitination) |
|
No change in mRNA/protein |
Functional changes |
Post-translational modifications or altered activity |
When qPCR and WB results conflict, avoid dismissing the data. Systematically troubleshoot technical errors and integrate multi-omics approaches (e.g., RNA-seq, proteomics) to uncover deeper regulatory mechanisms.
Remember: Great scientific questions often emerge from the cracks of "contradictions"!
